XNAT Open-Source Informatics for Imaging Research
XNAT is a web-based open source imaging informatics platform that facilitates common management, productivity, and quality assurance tasks for imaging and associated data. It can be used to support a wide range of imaging-based research projects.
XNAT
Originally developed by the Neuroinformatics Research Group at Washington University, XNAT is used by many research and clinical institutions all over the world.

XNAT has an extensible architecture that provides the capability of extending its functionality via plugins. XNAT out of the box can handle a wide range of imaging modalities (MRI, PET, Ultrasound, MEG, etc) and can easily be extended to capture any type of data using the plugin architecture.
XNAT uses a secure rest API for managing access to data on the server. This allows for customized workflows to be developed that bypass the XNAT server architecture itself and thus allow for rapid development and deployment of applications that utilize the main benefits of XNAT without the need to create a full-blown plugin.
Users will be able to store their deidentified data in XNAT and use metadata fields within XNAT to reference clinical data that is stored within UA Soteria. Leveraging the excellent extensibility of XNAT either through the plugin architecture or through REST API calls, users will be able to develop sophisticated pipelines and applications to analyze and explore their data.
Request access to get started using XNAT
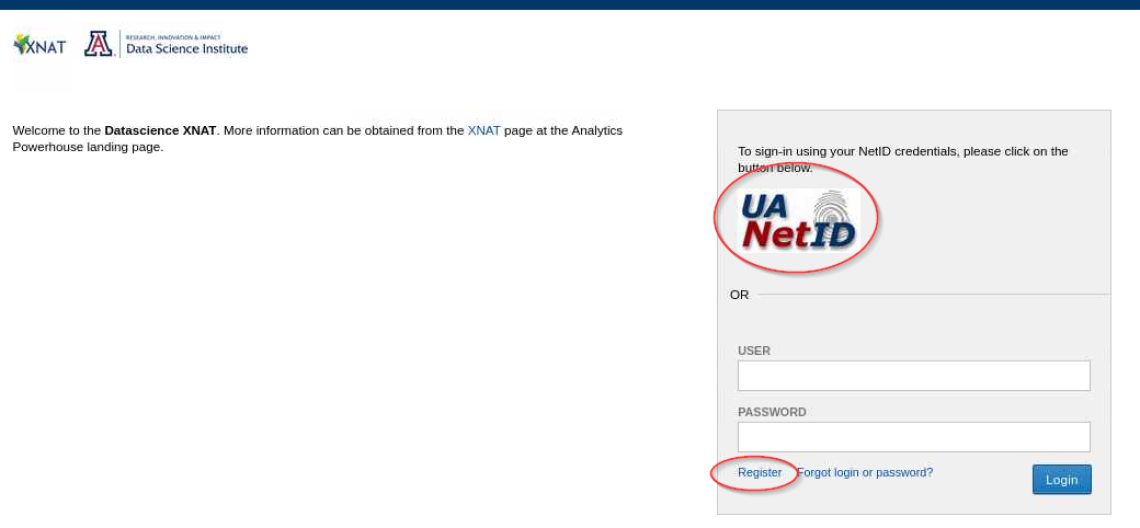
To request access to XNAT, please visit and click on the UA Net ID logo to automatically register at http://xnat.datascience.arizona.edu.
If you do not have a UA Net ID then click on the "Register" link to manually create a new user registration.
After registering, please wait for an email from the administrator to confirm that your access is enabled.
For more information on how XNAT can be used in your research, contact Chidi Ugonna at chidiugonna@arizona.edu.
FAQ's
XNAT is designed to manage the entire workflow of large imaging studies from the image ingress stage (typically from an imaging device like an MRI scanner) right up to quality control, adverse reporting, data analysis, data visualization and data sharing.
Users are granted access on a per-project basis and permissions available to users can be carefully tailored to protect the integrity of data and facilitate customized workflows.
Metadata is stored alongside imaging data allowing for sophisticated searching capabilities and for robust data curation and provenance. All actions performed on data are logged in an audit trail ensuring that all user actions performed on data can be tracked and traced accurately.
Yes. As an open-source application with a large and growing user base, the number and sophistication of plugins available to the community continues to grow and add value to XNAT.
Some notable plugins are the container service which allows users to run custom preprocessing pipelines on their data using container technology and the OHIF-Viewer which provides a state-of-the-art image viewer that allows DICOMS to be viewed using a web interface. There are several other plugins like LDAP support, Dicom Query Retrieve for querying PACS systems and XSYNC for syncing data between XNAT servers that are also useful.