OMERO is designed for Scientists
IMPORT DATA - With the desktop application (Windows, MacOS, Linux) or by command line. BIO-FORMATS directly handles the proprietary file formats on import and retains the relevant metadata.
ORGANIZE - View and organize your data from anywhere that you have internet access. In addition to imported metadata, labs can add additional information about the experiment, draw regions of interest, and annotate the images. The database is searchable with the ability to use keyword tagging.
VIEW - Work with your images using the OMERO.insight desktop app, with OMERO.web in your browser, or from 3rd party software.
ANALYZE - Use ImageJ, CellProfiler, QuPath, MATLAB, R, Python, scripts and other tools to run analysis on data in OMERO and save results on the server.
SHARE - Share data with collaborators or your PI using the Groups/Users permission system. The permissions system allows the PI to control who is part of their group and who has access to specific datasets.
PUBLISH - Publish direct from the server via a customized website or embedded viewer. Shared image data on an OMERO server can be configured to be complaint with the FAIR criteria ensuring that the data is findable, accessible, interoperable and reusable.
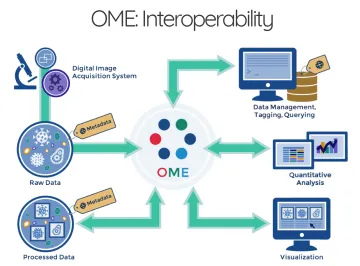
Why use OMERO?
With both of the major US funding agencies (NSF, NIH) now requiring a data management and sharing plan, University of Arizona microscope users are looking for ways to effectively store and make available their image data. There are specific repositories for other types of data (e.g., software, genomics, etc), but microscopy data can be very large (individual files and in aggregate). File sharing of image data can be a challenge, because not only are the data large, there are over 150 different proprietary image file formats. Supported by the University of Arizona Strategic Plan (UAHS 5.3 Health Analytics Powerhouse) the University of Arizona Data Science Institute is launching a local instance of the OMERO (Open Microscopy Environment Remote Objects) server for the storage, management and sharing of microscopy image data.
Appropriate data types for OMERO
Routine research image data (optical and electron microscopes, gel imagers, HCS, digital cameras, etc.) would be appropriate for OMERO. Human subjects-related data must be completely de-identified prior to uploading to the OMERO server.
How to get started
January 2023 - The OMERO project began it's first pilot project. Dr. Curiel's research project combined de-identified images taken in a clinic with clinical data securely stored in RedCap and biospecimen information stored in OpenSpecimen. Read more in the University of Arizona Health Science article, Revolutionizing skin cancer care through data.
August 2023 - A two-day, hands-on OMERO training workshop was held. Several new projects will come from the cohort of people who attended the workshop.
Getting on board - We anticipate needing to add laboratories in waves in 2023. This will allow for manageable sized training classes and allow time for the OMERO server to safely import what could be many terabytes of data
Contact us – We have a dedicated email address for this project. Reach out to us at RII-OMERO@arizona.edu.
OMERO project contact:
Doug Cromey, MS
Image Data Management Specialist, Data Science Institute, RII
Co-Manager, Imaging Cores-Optical, RII
(520) 626-2824
dcromey@arizona.edu